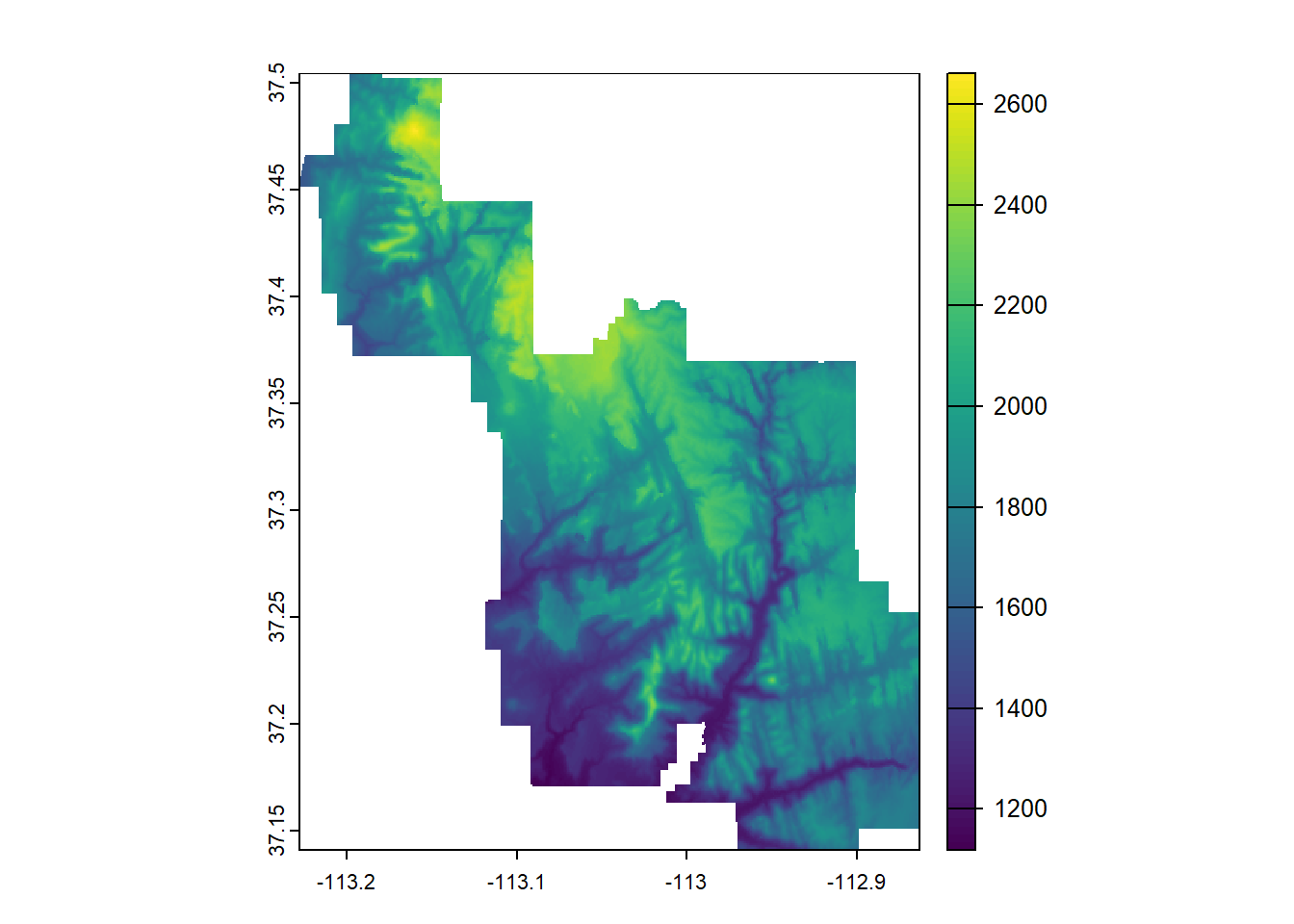
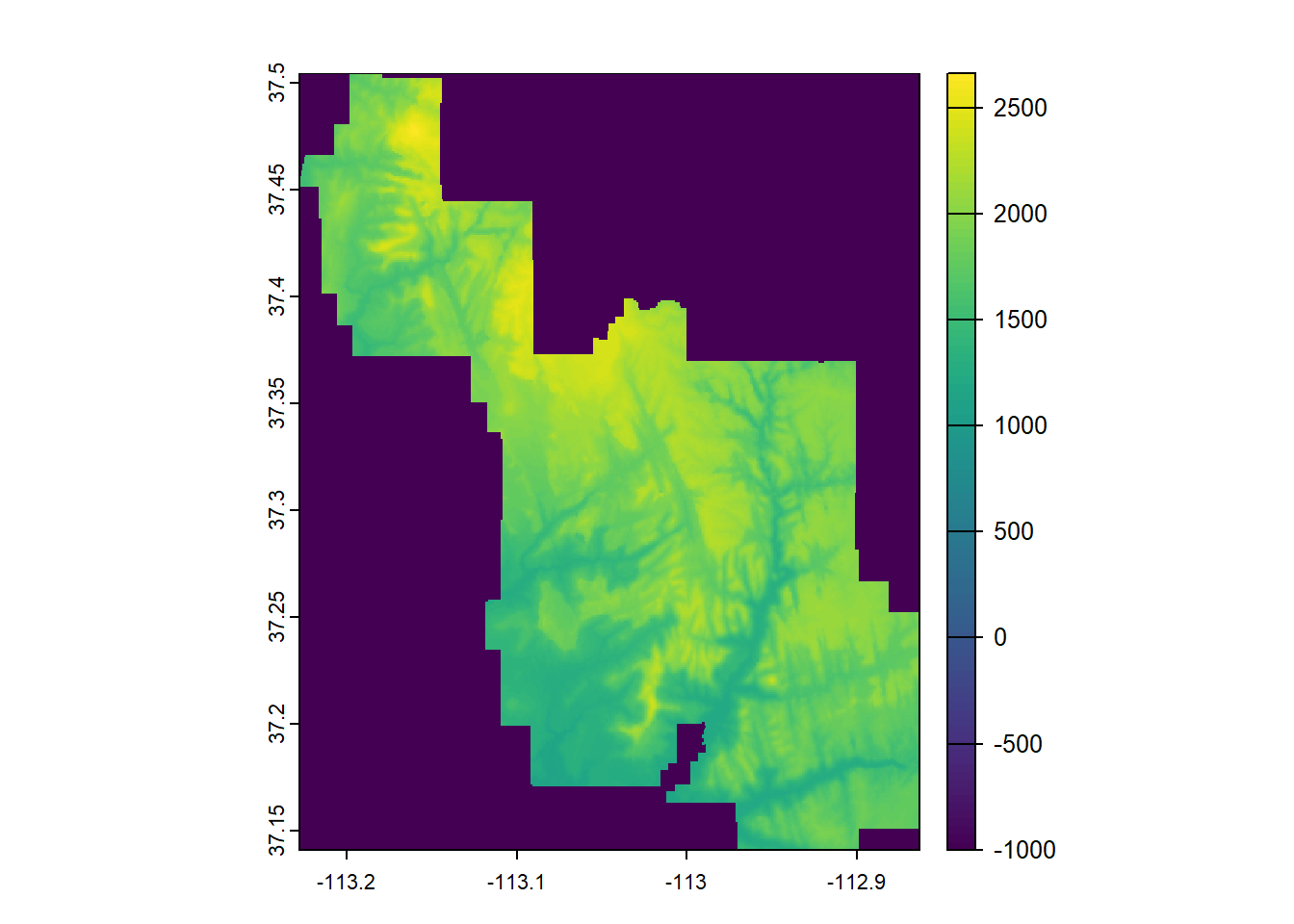
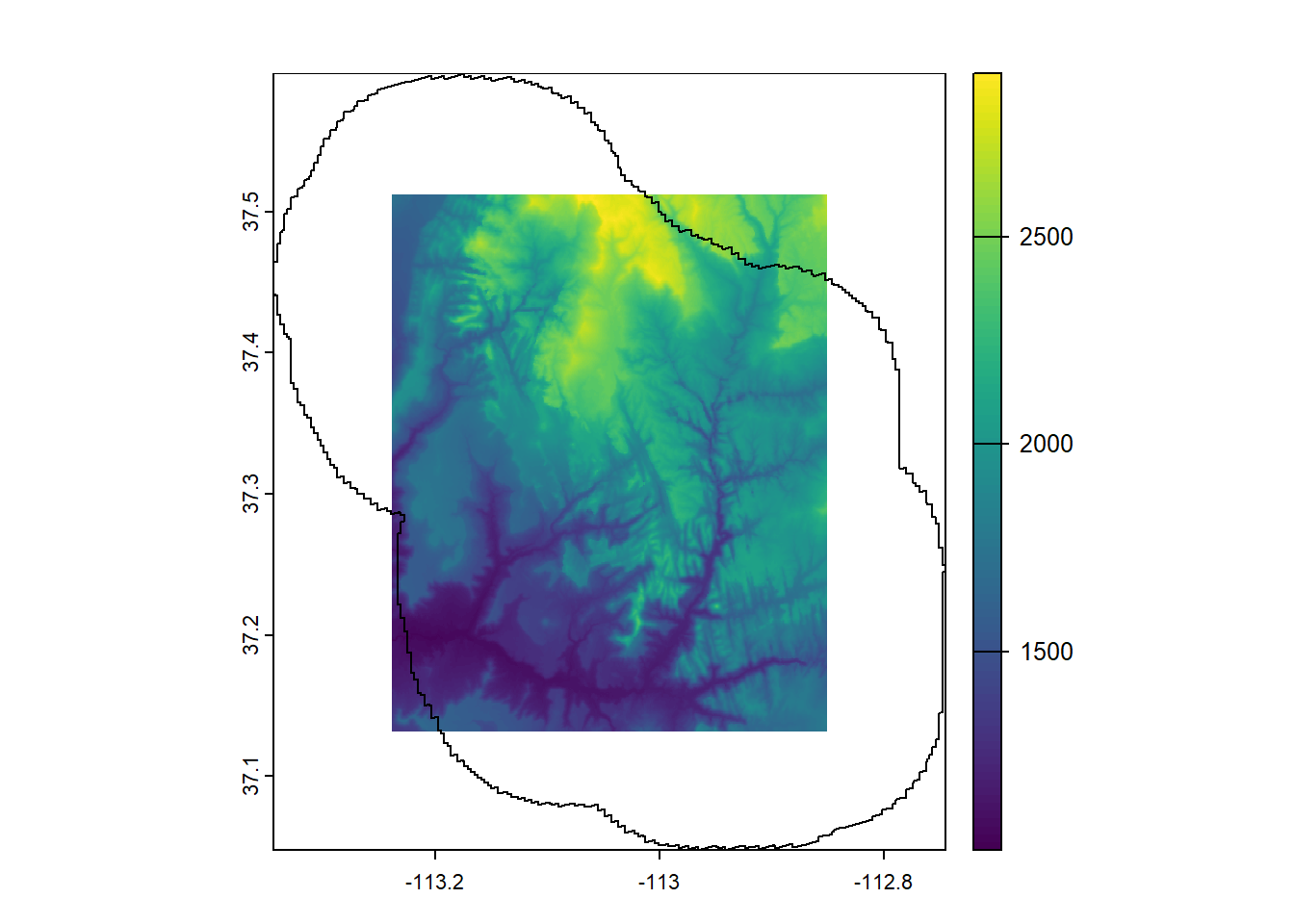
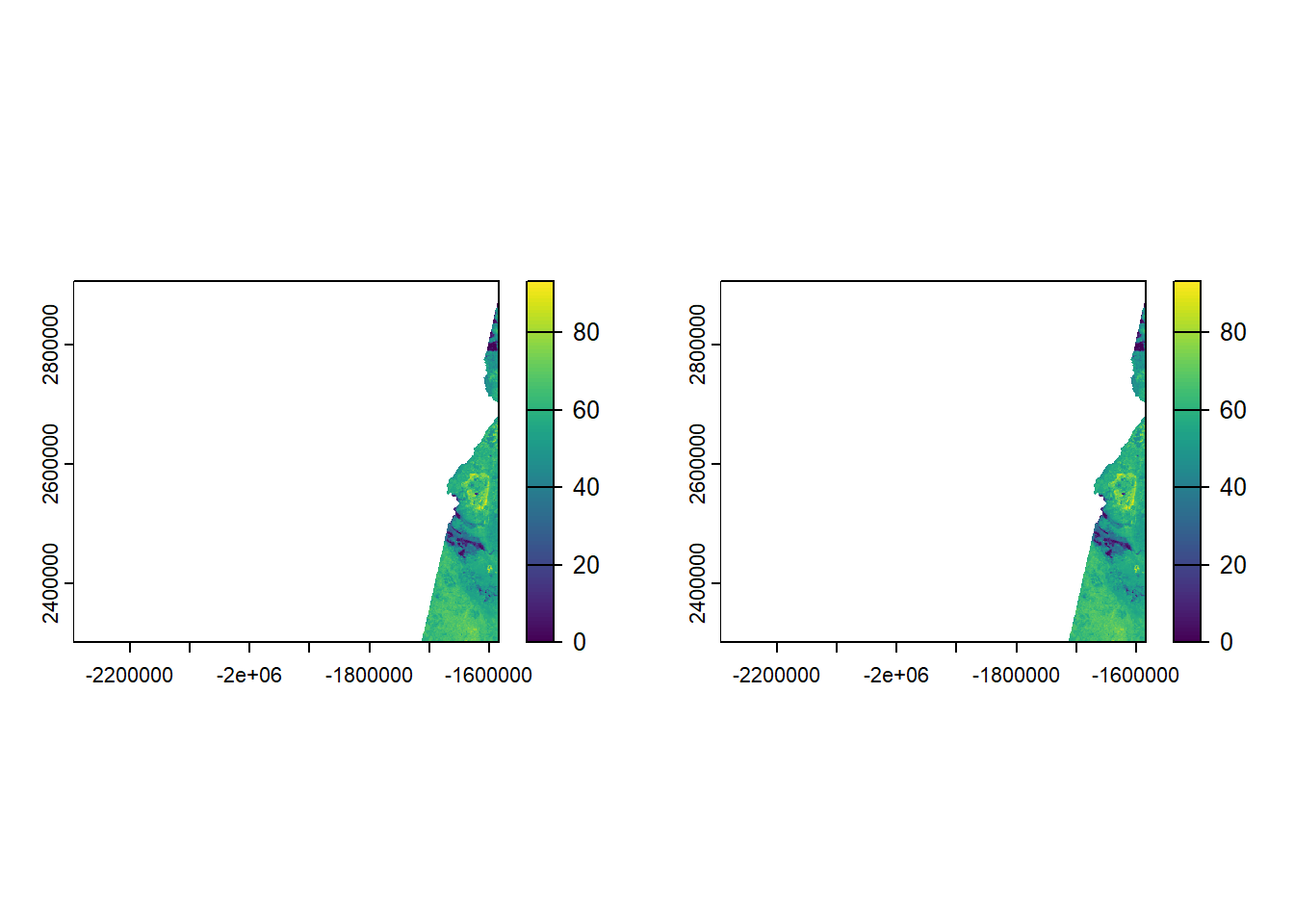
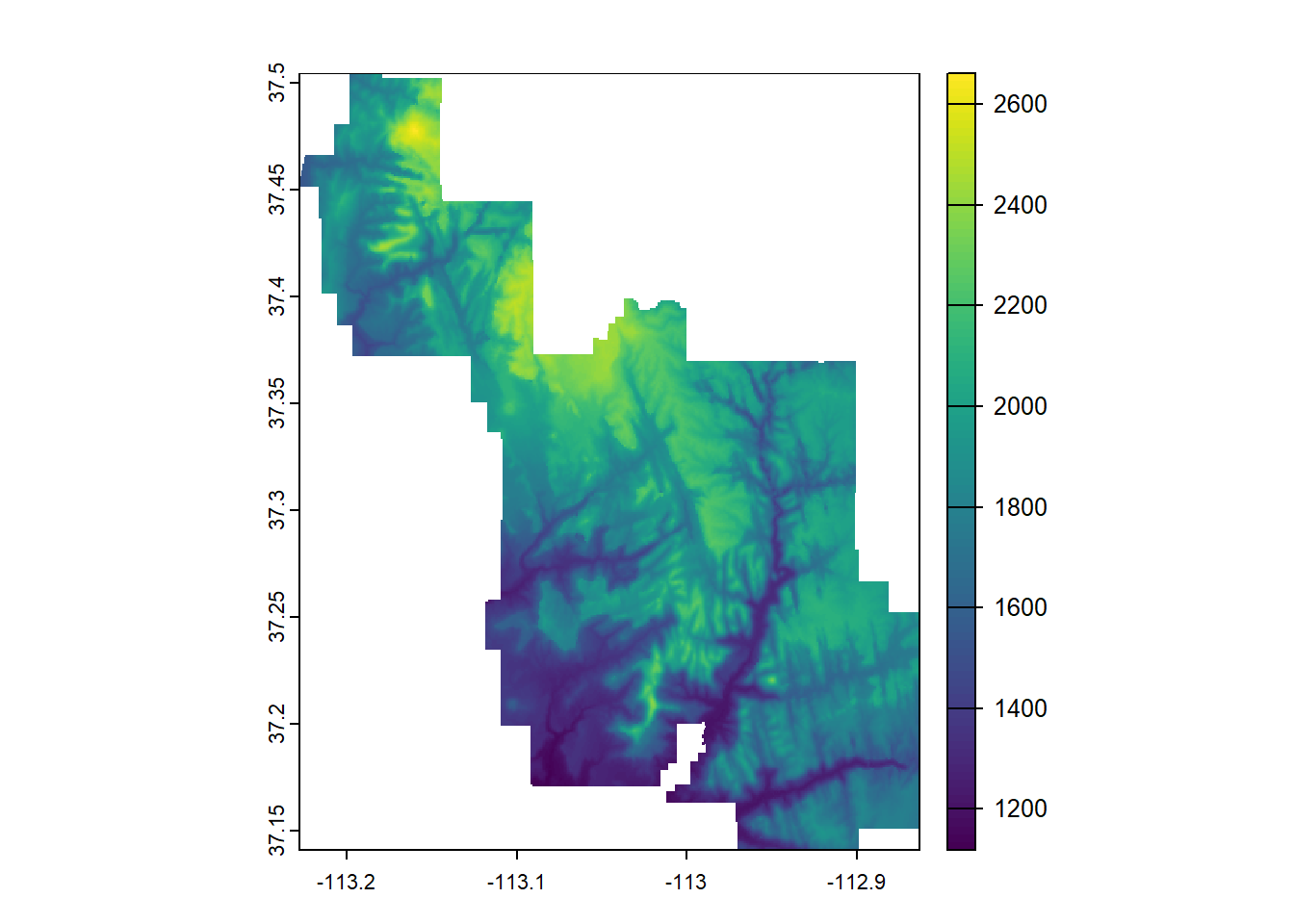
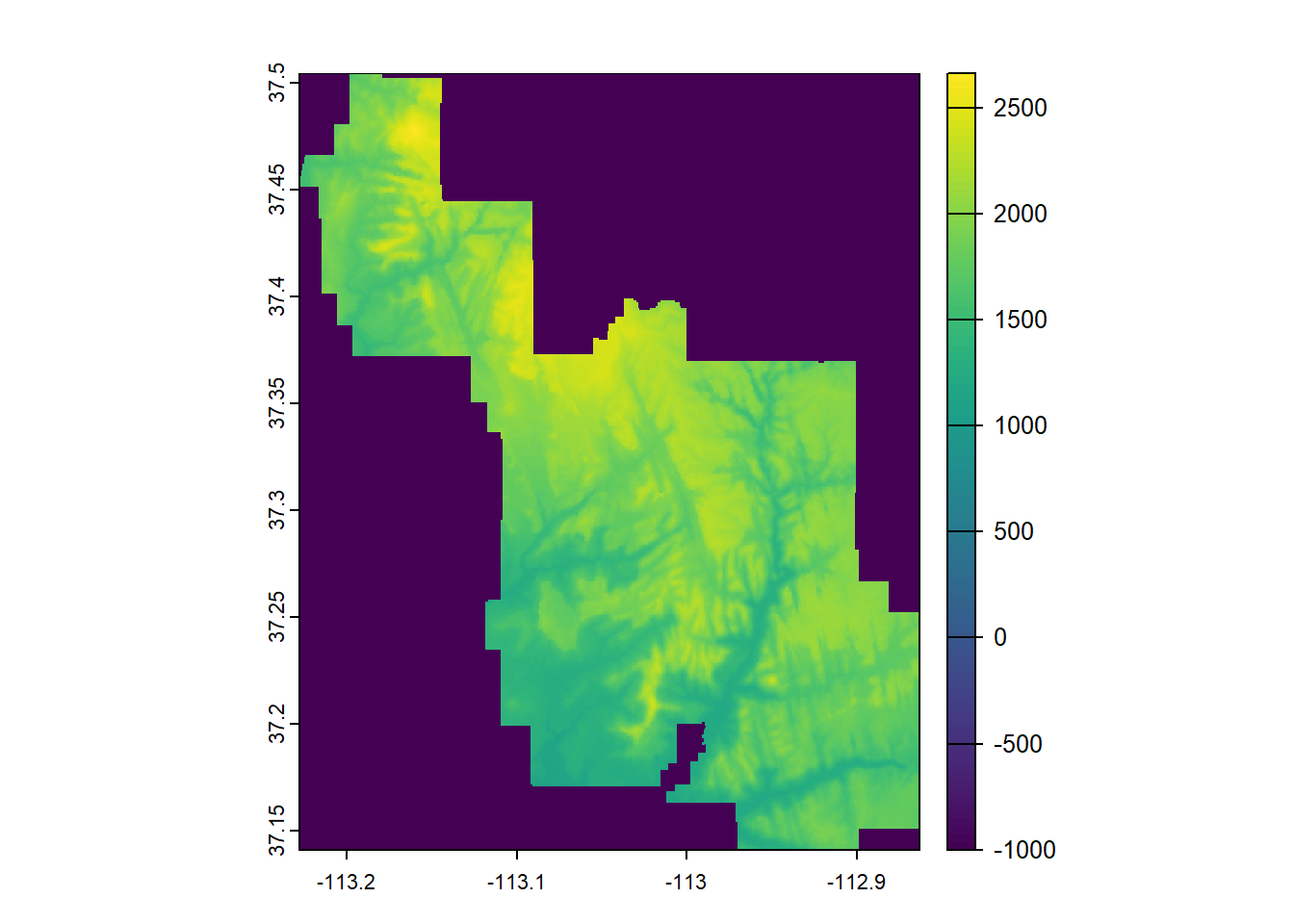
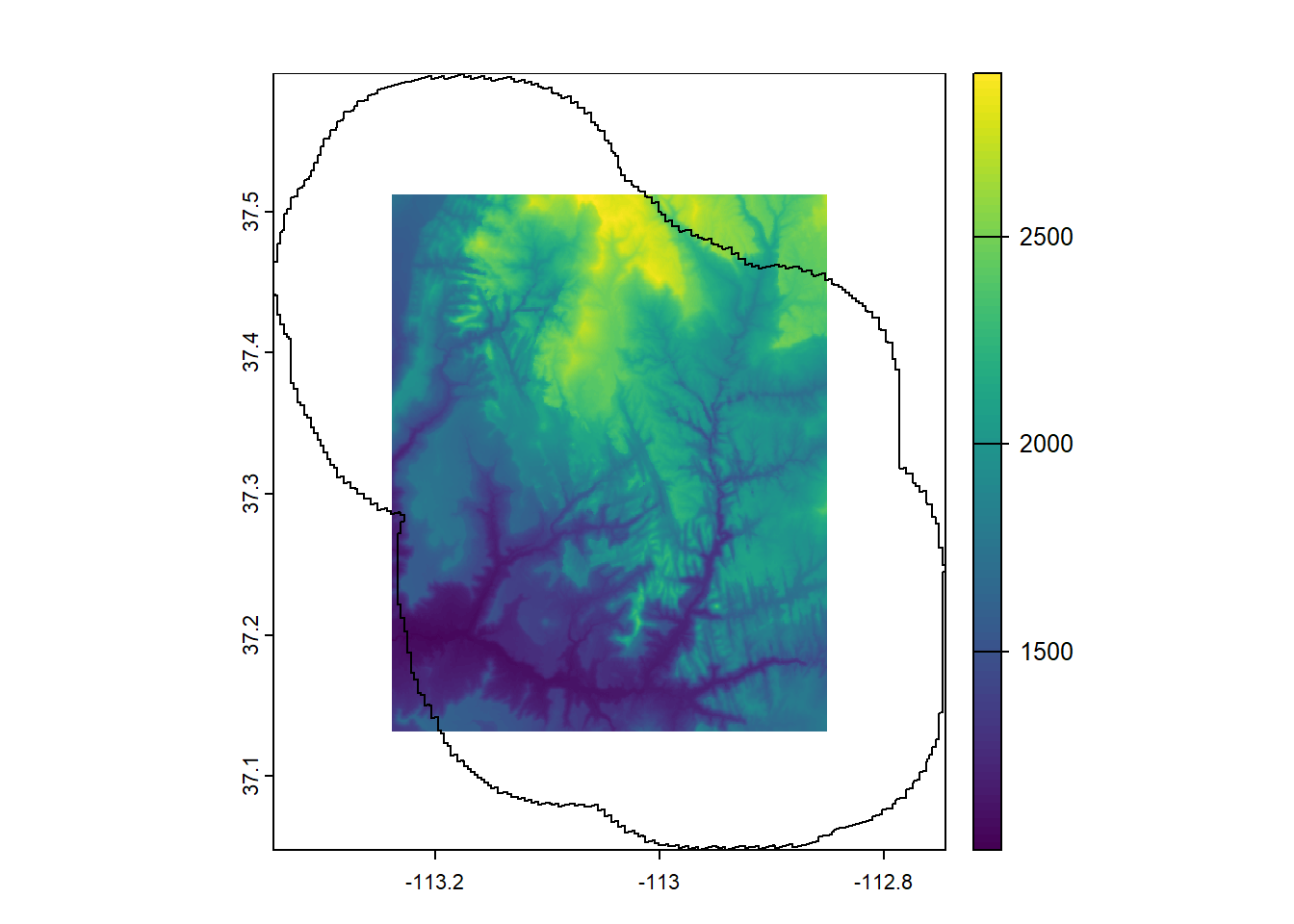
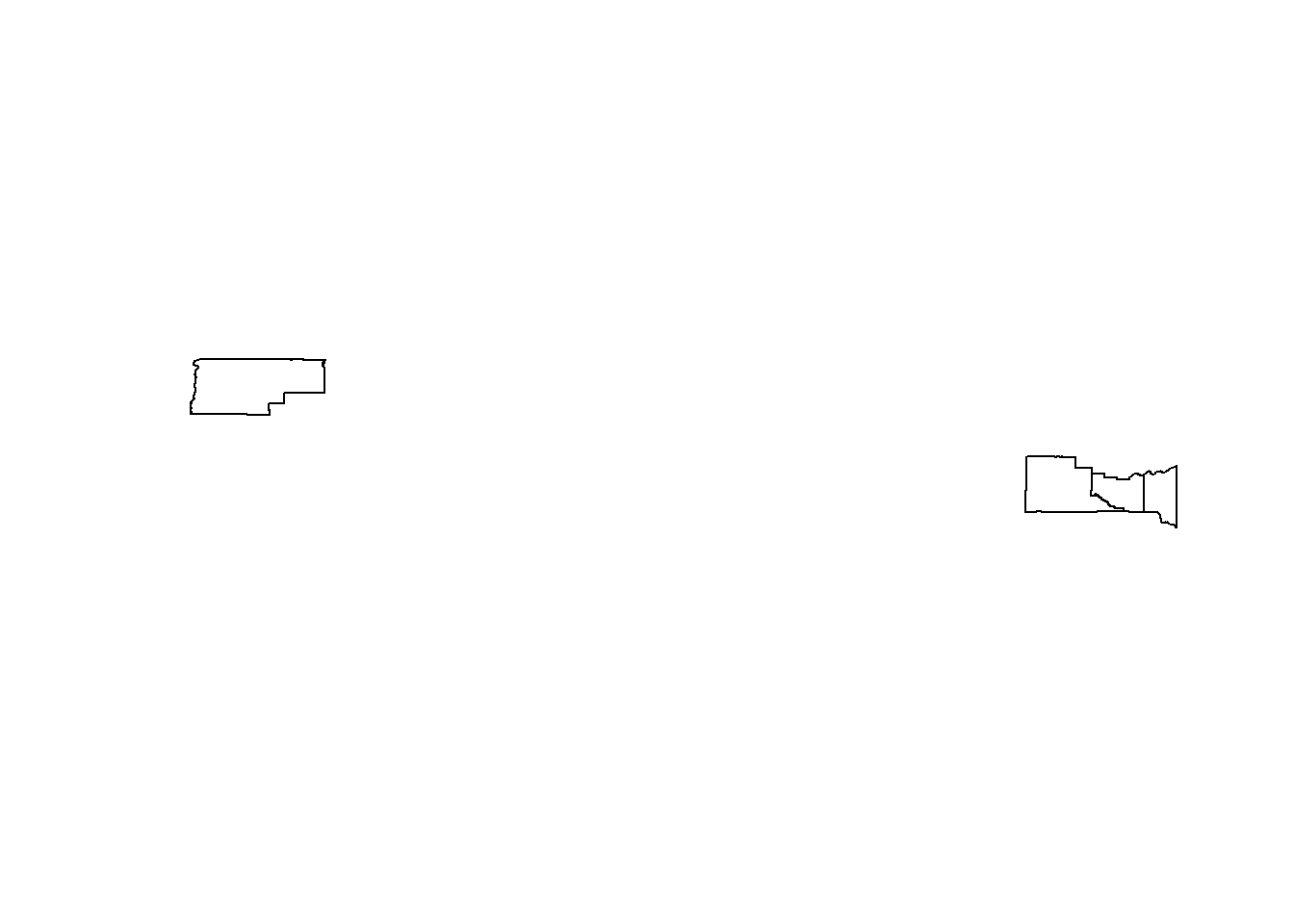--- title: "Session 12 Code" format: html --- ## Read in packages: ```{r} library (sf)library (terra)library (spDataLarge)library (tigris)library (tidyverse)``` ## Changing resolution: ```{r} <- rast ()``` ```{r} values (r) <- 1 : ncell (r)plot (r)``` ```{r} <- aggregate (r, 20 )``` ```{r} # rarely used <- disagg (r, 20 )plot (rd)``` ## Crop and Mask ```{r} = rast (system.file ("raster/srtm.tif" , package = "spDataLarge" ))= read_sf (system.file ("vector/zion.gpkg" , package = "spDataLarge" ))= st_transform (zion, crs (srtm))crs (srtm) == crs (zion)``` ```{r} <- crop (x= srtm, y= zion, snap= "near" )``` ```{r} <- crop (x= srtm, y= vect (zion), snap= "near" , mask= TRUE )plot (srtm.crop.msk)``` ```{r} <- mask (srtm.crop, vect (zion), updatevalue= - 1000 )plot (srtm.msk)``` ```{r} <- mask (srtm.crop, vect (zion), inverse= TRUE , updatevalue= - 1000 )``` ## Extend ```{r} <- zion %>% st_buffer (., 10000 )<- extend (srtm, vect (zion.buff))ext (srtm.ext)plot (srtm.ext)plot (st_geometry (zion.buff), add= TRUE )``` ## Practice ```{r} #| eval: false <- rast ("/opt/data/data/rasterexample/Copy of CRPS_ID.tif" )<- rast ("/opt/data/data/rasterexample/Copy of CRPS_OR.tif" )``` ```{r} #| include: false <- rast ("C:/Users/carolynkoehn/Documents/HES505_Fall_2024/data/2023/rasterexample/Copy of CRPS_ID.tif" )<- rast ("C:/Users/carolynkoehn/Documents/HES505_Fall_2024/data/2023/rasterexample/Copy of CRPS_OR.tif" )``` ### Aggregate ```{r} <- aggregate (id, fact = 30 )<- aggregate (or, fact = 30 )``` ### Check for alignment ```{r} crs (id_agg) == crs (or_agg)origin (id_agg) == origin (or_agg)ext (id_agg) == ext (or_agg)``` ### Align the origins `project` and `resample` don't give the intended results.```{r} <- project (id_agg, or_agg)<- resample (id_agg, or_agg)par (mfrow = c (1 ,2 ))plot (id_proj)plot (id_resamp)``` ```{r} <- extend (id_agg, or_agg)<- extend (or_agg, id_ext)<- resample (id_ext, or_ext)``` ```{r} crs (id_resamp) == crs (or_ext)origin (id_resamp) == origin (or_ext)ext (id_resamp) == ext (or_ext)``` ### Mosaic ```{r} <- mosaic (id_resamp, or_ext)``` ### Crops and Masks ```{r} <- counties (state = c ("ID" , "OR" ))<- idor_counties %>% filter (NAME %in% c ("Teton" , "Jefferson" , "Madison" ))plot (st_geometry (east_id))``` `mask` without `crop` leaves a lot of white space.```{r} <- st_transform (east_id, crs (idor))<- mask (x = idor, mask = east_id_proj)<- crop (idor, east_id_proj, mask= TRUE )``` ## Read in packages: ```{r} library (sf)library (terra)library (spDataLarge)library (tigris)library (tidyverse)``` ## Changing resolution: ```{r} <- rast ()``` ```{r} values (r) <- 1 : ncell (r)plot (r)``` ```{r} <- aggregate (r, 20 )``` ```{r} # rarely used <- disagg (r, 20 )plot (rd)``` ## Crop and Mask ```{r} = rast (system.file ("raster/srtm.tif" , package = "spDataLarge" ))= read_sf (system.file ("vector/zion.gpkg" , package = "spDataLarge" ))= st_transform (zion, crs (srtm))crs (srtm) == crs (zion)``` ```{r} <- crop (x= srtm, y= zion, snap= "near" )``` ```{r} <- crop (x= srtm, y= vect (zion), snap= "near" , mask= TRUE )plot (srtm.crop.msk)``` ```{r} <- mask (srtm.crop, vect (zion), updatevalue= - 1000 )plot (srtm.msk)``` ```{r} <- mask (srtm.crop, vect (zion), inverse= TRUE , updatevalue= - 1000 )``` ## Extend ```{r} <- zion %>% st_buffer (., 10000 )<- extend (srtm, vect (zion.buff))ext (srtm.ext)plot (srtm.ext)plot (st_geometry (zion.buff), add= TRUE )``` ## Practice ```{r} #| eval: false <- rast ("/opt/data/data/rasterexample/Copy of CRPS_ID.tif" )<- rast ("/opt/data/data/rasterexample/Copy of CRPS_OR.tif" )``` ```{r} #| include: false <- rast ("C:/Users/carolynkoehn/Documents/HES505_Fall_2024/data/2023/rasterexample/Copy of CRPS_ID.tif" )<- rast ("C:/Users/carolynkoehn/Documents/HES505_Fall_2024/data/2023/rasterexample/Copy of CRPS_OR.tif" )``` ### Aggregate ```{r} <- aggregate (id, fact = 30 )<- aggregate (or, fact = 30 )``` ### Check for alignment ```{r} crs (id_agg) == crs (or_agg)origin (id_agg) == origin (or_agg)ext (id_agg) == ext (or_agg)``` ### Align the origins `project` and `resample` don't give the intended results.```{r} <- project (id_agg, or_agg)<- resample (id_agg, or_agg)par (mfrow = c (1 ,2 ))plot (id_proj)plot (id_resamp)``` ```{r} <- extend (id_agg, or_agg)<- extend (or_agg, id_ext)<- resample (id_ext, or_ext)``` ```{r} crs (id_resamp) == crs (or_ext)origin (id_resamp) == origin (or_ext)ext (id_resamp) == ext (or_ext)``` ### Mosaic ```{r} <- mosaic (id_resamp, or_ext)``` ### Crops and Masks ```{r} <- counties (state = c ("ID" , "OR" ))<- idor_counties %>% filter (NAME %in% c ("Teton" , "Jefferson" , "Madison" ))plot (st_geometry (east_id))``` `mask` without `crop` leaves a lot of white space.```{r} <- st_transform (east_id, crs (idor))<- mask (x = idor, mask = east_id_proj)<- crop (idor, east_id_proj, mask= TRUE )```