Code
library(terra)
library(spDataLarge)
library(sf)Carolyn Koehn
Load libraries:
Get land cover data:
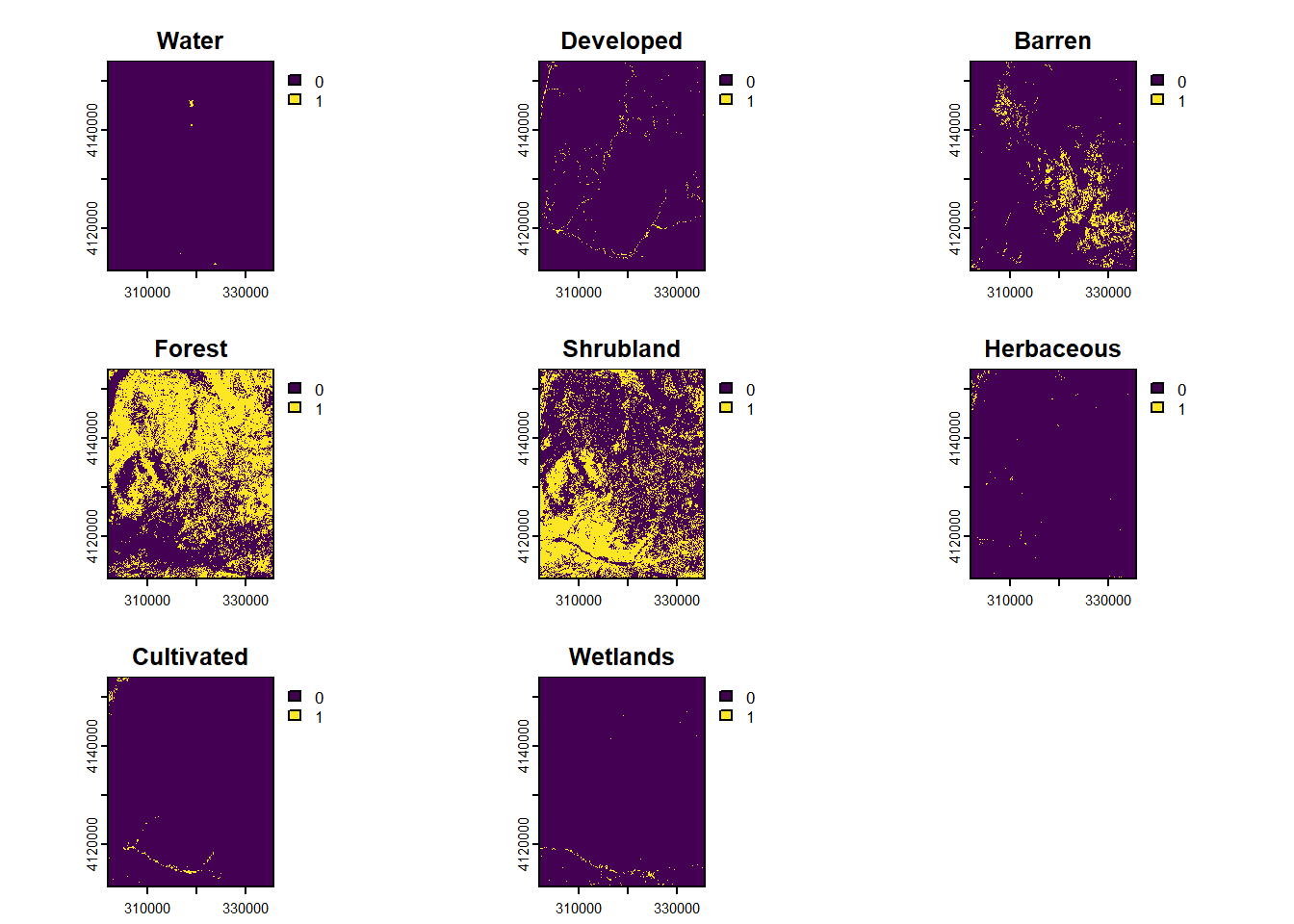
Separate categorical raster into Boolean layers:
Get slope data:
Prepare overlay layers:
Run overlay analysis:
Get data:
# get presence-absence simulated data
presabs <- st_read("/opt/data/data/presabsexample/presenceabsence.shp", quiet = TRUE)
# get predictor data file paths
preds.list <- list.files("/opt/data/data/presabsexample", "grd$", full.names = TRUE)
# get predictors as raster stack
pred.stack <- rast(preds.list)
# rename layers of raster stack
names(pred.stack) <- c("MeanAnnTemp", "TotalPrecip", "PrecipWetQuarter", "PrecipDryQuarter", "MinTempCold", "TempRange")Extract predictor data at each point and add presence/absence column called y:
Fit logistic regression model:
Call:
glm(formula = y ~ TempRange + PrecipDryQuarter, family = binomial(link = "logit"),
data = pts.df)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.2820 0.3789 -3.384 0.000715 ***
TempRange -3.5156 0.7984 -4.403 1.07e-05 ***
PrecipDryQuarter 0.4157 0.5938 0.700 0.483969
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 125.37 on 99 degrees of freedom
Residual deviance: 59.18 on 97 degrees of freedom
AIC: 65.18
Number of Fisher Scoring iterations: 6Predict model results across entire study area:
---
title: "Session 21 code"
author: "Carolyn Koehn"
format: html
---
Load libraries:
```{r}
#| message: false
#| warning: false
library(terra)
library(spDataLarge)
library(sf)
```
## Overlays
Get land cover data:
```{r}
nlcd <- rast(system.file("raster/nlcd.tif", package = "spDataLarge"))
plot(nlcd)
```
Separate categorical raster into Boolean layers:
```{r}
nlcd.segments <- segregate(nlcd)
# rename layers of raster stack
names(nlcd.segments) <- levels(nlcd)[[1]][-1,2]
plot(nlcd.segments)
```
Get slope data:
```{r}
# elevation data
srtm <- rast(system.file("raster/srtm.tif", package = "spDataLarge"))
# get slope
slope <- terrain(srtm, v = "slope")
```
Prepare overlay layers:
```{r}
# only slopes less than 10 are suitable
suit.slope <- slope < 10
# only shrubland is suitable
suit.landcov <- nlcd.segments["Shrubland"]
# make sure raster layers align
suit.slope.match <- project(suit.slope, suit.landcov)
```
Run overlay analysis:
```{r}
suit <- suit.slope.match + suit.landcov
plot(suit)
```
## Logistic regression
Get data:
```{r}
#| eval: false
# get presence-absence simulated data
presabs <- st_read("/opt/data/data/presabsexample/presenceabsence.shp", quiet = TRUE)
# get predictor data file paths
preds.list <- list.files("/opt/data/data/presabsexample", "grd$", full.names = TRUE)
# get predictors as raster stack
pred.stack <- rast(preds.list)
# rename layers of raster stack
names(pred.stack) <- c("MeanAnnTemp", "TotalPrecip", "PrecipWetQuarter", "PrecipDryQuarter", "MinTempCold", "TempRange")
```
```{r}
#| include: false
# get presence-absence simulated data
presabs <- st_read("C:/Users/carolynkoehn/Documents/HES505_Fall_2024/data/2023/session28/session28/presenceabsence.shp", quiet = TRUE)
# get predictor data file paths
preds.list <- list.files("C:/Users/carolynkoehn/Documents/HES505_Fall_2024/data/2023/session28/session28", "grd$", full.names = TRUE)
# get predictors as raster stack
pred.stack <- rast(preds.list)
# rename layers of raster stack
names(pred.stack) <- c("MeanAnnTemp", "TotalPrecip", "PrecipWetQuarter", "PrecipDryQuarter", "MinTempCold", "TempRange")
```
Extract predictor data at each point and add presence/absence column called `y`:
```{r}
pts.df <- terra::extract(pred.stack, presabs)
pts.df[,2:7] <- scale(pts.df[,2:7])
pts.df$y <- presabs$y
```
Fit logistic regression model:
```{r}
logistic.carolyn <- glm(y ~ TempRange + PrecipDryQuarter,
family = binomial(link = "logit"),
data = pts.df)
summary(logistic.carolyn)
```
Predict model results across entire study area:
```{r}
newpreds <- predict(pred.stack, logistic.carolyn, type = "response")
plot(newpreds)
```